AMPure XP Beads for DNA Cleanup
Applications: PCR cleanup, DNA sample concentration, NGS library prep
The gold standard in bead-based, next-generation sequencing (NGS) cleanup—in fact, it's suggested in over 200 library preparation kits from trusted sequencing companies.
Order AMPure XP Reagent Right Away
Explore PCR Models
We're Better Together: Become an OEM Partner. Learn More →
Overview
The AMPure XP bead-based reagent is intended for performing DNA cleanup steps in different genomic applications like sequencing, qPCR/ddPCR/PCR, microarrays, and other enzymatic reactions. It is powered by proven SPRI technology, which uses paramagnetic beads to selectively bind nucleic acids by type and size.
![]()
Works with DNA
![]()
High recovery of amplicons, greater than 100 bp
![]()
Efficient removal of unincorporated dNTPs, primers, primer dimers, salts and other contaminants
![]()
Predictable and consistent size selection
Maximizing recovery, consistency, and speed to facilitate the entire NGS workflow, the AMPure XP reagent meets the stringent needs of today’s genomic applications and minimizes the risk of losing important genetic information. That's why it's suggested in over 200 NGS library preparation kits often as the sole cleanup option, including those from the industry's most trusted sequencing companies like Illumina, Oxford Nanopore Technologies, PacBio and Thermo Scientific.
Workflows and Protocol
AMPure XP Manual Workflow
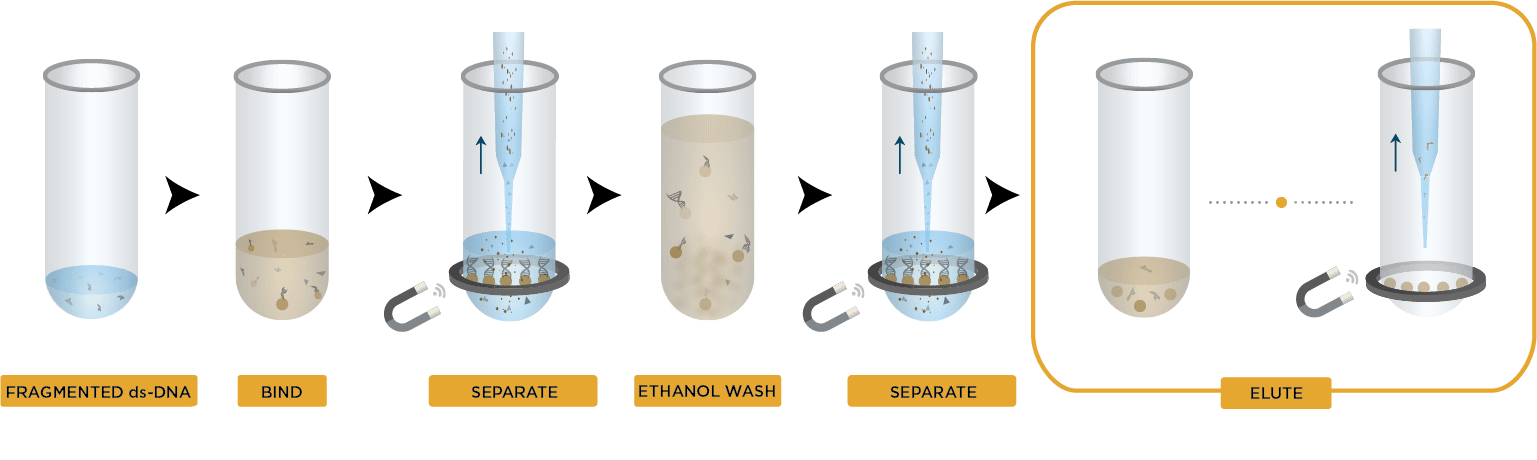
- Bind DNA to magnetic beads
- Separate beads from contaminants
- Wash magnetic beads with 70% ethanol to remove contaminants
- Elute DNA from magnetic beads
- Transfer to new plate
AMPure XP bead-based reagent can be performed either manually or automated on a liquid handling system depending on batch size or overall throughput.
In the table you can see estimated hands-on time and total time, in minutes, required to perform 48, 96, and 4 x 96 AMPure XP cleanups.
To learn more about automation of the AMPure XP PCR Purification system on the Biomek i5 Multichannel 96 Genomics Workstation, read our application note.
| Manual Workflow | Automated Workflow | |||
|---|---|---|---|---|
| Batch Size | 48 | Hands-on Time | 25 | 5 |
| Total Time | 25 | 22 | ||
| 96 | Hands-on Time | 30 | 5 | |
| Total Time | 30 | 22 | ||
| 4 x 96 | Hands-on time | NR | 10 | |
| Total Time | NR | 46 | ||
NR = Not Recommended
Specifications
| Nucleic Acid Input | PCR products, fragmented DNA |
| Output | DNA |
| Recovery | AMPure XP reagent was developed for the purification of PCR products 100bp<. Normal recovery ranges from 60-90%, and is typically between 70-90%. |
| Bead Ratio | 1.8X for cleanup. Learn more about bead size selection and why the ratio matters. |
| Format | Liquid |
| Volumes | 5 mL, 60 mL, or 450 mL |
| Applications | PCR Purification, DNA Cleanup, NGS Cleanup |
| Processing mode | Automated or manual |
| Technology | SPRI paramagnetic bead-based technology |
| Storage | 4 °C |
How to Automate AMPure XP DNA Cleanup
When compared to manual operations, the AMPure XP beads automated on Biomek liquid handlers provides:
- Reduced hands-on time and increased throughput
- Option to run the method end-to-end with only setup and tear-down touch points
- Reduction in pipetting errors
- Standardized workflow for improved results
- Quick implementation with demonstrated methods
To learn more about automation of the AMPure XP beads on the Biomek i5 Nucleic Acid Cleanup Solution, read our method overview.
To discuss the feasibility to automate the AMPure XP DNA cleanup in your lab with one of our experts complete this form.

Frequently Asked Questions
The reagent will bind DNA and RNA; however, the RNAClean XP reagent is manufactured under RNase-free conditions and is QC tested to be RNase free whereas the AMPure XP reagent is not.
The AMPure XP reagent is manufactured and tested for the storage temperature indicated on the bottle and we can guarantee performance only at that temperature.
Find Beckman Coulter Life Sciences AMPure XP technical documents here.
Although size selection protocols developed by other organizations do exist, we cannot guarantee performance or support this application. We suggest using the SPRIselect reagent, as it is developed specifically for size selection purposes. Want to learn more about the difference between the AMPure XP and SPRIselect reagents? Visit this page.
No. Any beads carried over to the final destination plate are inert in downstream enzymatic reactions.
Yes, but the recovery will decrease with a decrease in elution volume. In addition, if the beads cannot reach the magnet during the separation it may be necessary to leave a portion of the eluate behind if bead carryover must be avoided. Be sure to resuspend the beads fully in the elution buffer.
The binding capacity of the beads is so high that they cannot be exceeded with any sample type. It is possible to bind at least 7 μg of nucleic acid to 1 μL AMPure XP reagent, however the sample will become so viscous that it is difficult to pipette the sample.
The recovery depends on the fragment size, sample volume, sample concentration, elution volume and some other factors. Under ideal conditions recovery can be up to 90%, but under sub-optimal it can be as low as 60%.
Our beads rely upon a multifactorial equilibrium chemistry for binding, washing, and elution. Binding buffer components facilitate nucleic acid immobilization at functional-group-rich binding sites on the beads and the wash steps preserve this delicate equilibrium while solubilizing contaminants to allow for their removal. Elution disrupts this balance and re-solubilizes nucleic acids to allow for their separation from the beads.
A dry step can be incorporated after the removal of the last ethanol wash and before the addition of the elution buffer. For purification of very large fragments, such as gDNA, a dry step is not recommended because recovery would be decreased. Fragments up to 40 kb have been known to tolerate a dry step.
Didn't find an answer to your question? Talk to our expert.
Citations
AMPure XP reagents have been cited in over 52,000 publications on Google Scholar and referenced in articles in Science, Nature, and PNAS. Here are two featured articles:
Greenwald WW et al. Subtle changes in chromatin loop contact propensity are associated with differential gene regulation and expression. Nat Commun. 2019 Mar 5;10(1):1054. doi: 10.1038/s41467-019-08940-5.
Peffers MJ et al. Transcriptomic profiling of cartilage ageing. Genom Data. 2014 Mar 19;2:27-8. doi: 10.1016/j.gdata.2014.03.001.
Technische Dokumente
Products and demonstrated applications are not intended or validated for use in diagnostic procedures.